SimStack#
Simstack simplifies the developing and analysis of intricate simulation workflows, putting rapid prototyping with easy reach for research workers. Simstack is a simple-to-use GUI that allows users to set up jobs easily and do templated tasks. It considerably reduces the time required between making a project and running simulation, without requiring complex programming skills at every step of problem solving.
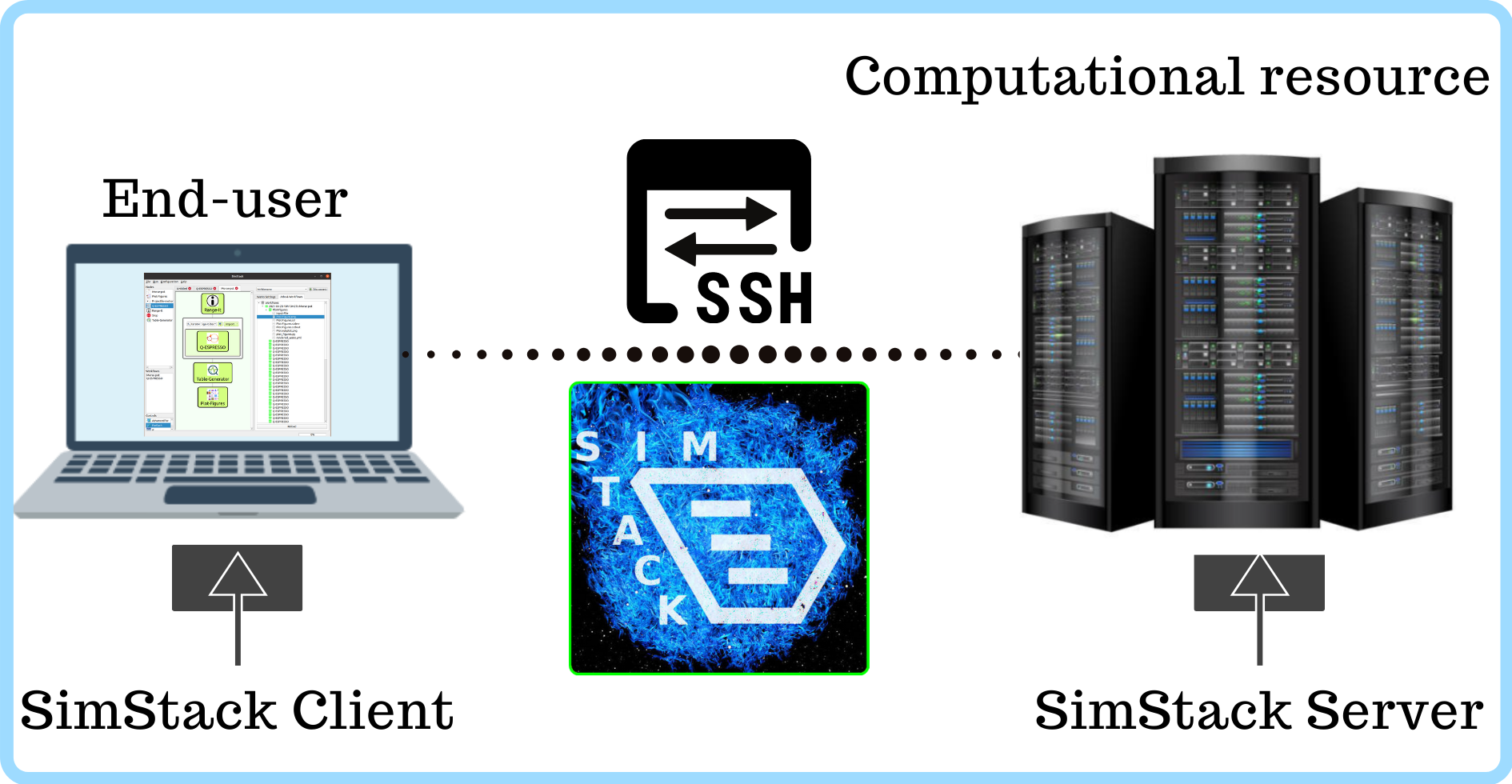
Simstack provides robust solutions for communication and management of HPC systems. It handles data transfer between phases of the workflow, job submissions and in particular takes care of opportunities with large amounts of information to be processed efficiently.
In essence, Simstack has one purpose and that is to simplify the job of computational research. By making advanced simulations possible and more understandable to carry out concurrently, more researchers have access to scientific experimentation.
Installation / Setup#
For SimStack installation and setup, we use Conda packages directly for its client and server components. When the server is started, it runs in the user space making auto processing possible once a user has finished interacting with it. Data exchange between client and server is carried out in SimStack by SSH, that safe and efficient. This simple approach reduces the complexity of the setup process.
Implementation of a new simulation code#
This section includes a description of the implementation of a new simulation code, which was written to set up a structure for calculations in Quantum Espresso. Thus, below presents a Python code with an XML configuration, for example. The atomic steps to create a structure are set up. In other words, step 1 is input and output data, as well as user-accessible parameters. In this simulation, the latter include a species of the elements, the crystal lattice parameters, and the k-point grid dimensions.
One of the main part of a SimStack workflow is an XML configuration file that describes the structure of the user inputs, the commands to run, and the input/output files needed for a simulation. Additionally, by integrating Jinja templating within the XML configuration, SimStack introduces a layer of dynamic parameter substitution, enhancing the flexibility and customization of simulations:
<WaNoTemplate>
<!-- Meta Information -->
<WaNoRoot name="Structure_creation">
<!-- Jinja Templating for User-defined Parameters -->
<WaNoString name="element">{{ wano.element }}</WaNoString>
<WaNoString name="a">{{ wano.a }}</WaNoString>
<WaNoBool name="cubic">{{ wano.cubic }}</WaNoBool>
</WaNoRoot>
<WaNoExecCommand>
<!-- Environment Setup and Script Execution -->
</WaNoExecCommand>
<!-- Input and Output File Definitions -->
</WaNoTemplate>
And here’s how it looks directly in SimStack:
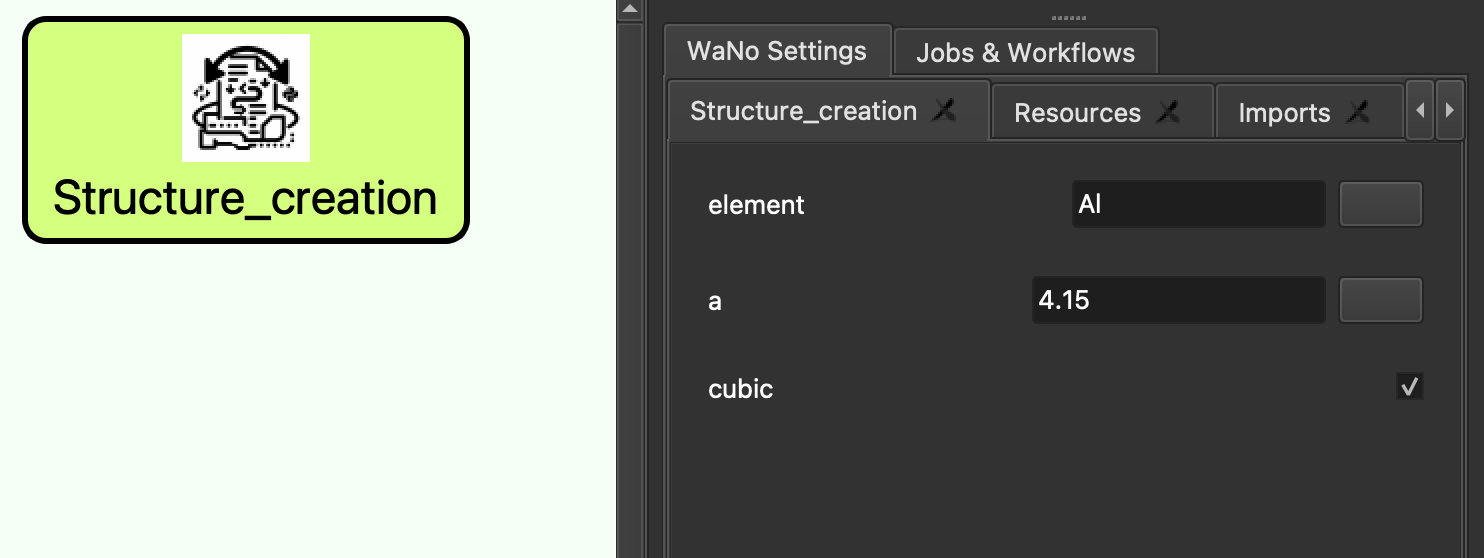
After starting the workflow, rendered_wano.yml file is produced by SimStack, which is all the parameters written by the user in XML configuration but in a form that allows to bridge the user input data and Python script which are all the parameter values, thus the simulation is executed with parameters exactly the same as the one the researcher specified.
The Python script first reads the rendered_wano.yml file and extracts the user-defined parameters. It is then used to create a structure file - structure.xyz by leveraging the ASE library:
import yaml
from ase.build import bulk
from ase.io import write
if __name__ == '__main__':
with open('rendered_wano.yml') as fh:
params = yaml.safe_load(fh)
element = params.pop('element')
struct = bulk(element, **params)
write("structure.xyz", struct)
This concludes the simulation that covers several key aspects: from defining atomic steps and parameters to integrating with HPC systems. The importance of this approach is emphasized by the need to ensure flexibility and modularity in the simulation creation process, achieved through the use of the Jinja templating engine for dynamic substitution of user-defined parameters and Python scripts for executing simulations.
Submission to an HPC / Check pointing / Error handling#
The migration of configurations is simple with Simstack, moving simulations from destined for a local system to the HPC. Users first run the simulation locally. Then safely, the simulation code and its inputs are moved to an HPC using SSH.
Simstack’s central role in mediating between the workflow and the scheduling system of the HPC begins once it is installed on an HPC. It continuously watches over your simulation, making sure each step proceeds as planned. When a step is completed, Simstack subsequently automates the next workflow step via its output (so long as this must be another job). This efficient setup minimizes all unnecessary waits and maximizes HPC resource use.
Data Storage / Data Sharing#
In our HPC systems, we designed a system to achieve data storage and re-use of computational data. The system employs hashes generated from workflow annotations (wano) and input parameters to pinpoint data throughout multiple generations of computation. The system efficiently identifies and retrieves data from previous work for reuse in later stages.
Simstack uses the file system as a base, it adds SQLite for indexing and retrieval. This approach eliminates wasted data replication, especially with huge data such as multi-terabyte trajectory datasets. It suggests the principle of conditional copying or creating a link to the actual data. Using these unique hashes to query a SQLite database, the system looks for previous computations with matching parameters. This method saves a significant amount of time and resource.
Publication of the workflow#
In the process of publishing a workflow, elements are organized into modules and configurations. These items are kept apart in order to maintain clarity and make the system easier to manage.
Inside the project directory, modules are stored as subfolders. This makes it possible for each component to be developed, tested and versioned independently. The modular approach makes use of Git submodules for handling dependencies and changes, resulting in a consistent workflow development that is stable.
The workflow, All module and configuration work is now ready for publishing. It gets uploaded or registered with a workflow-store repository set up specifically for the sharing of scientific workflows.
